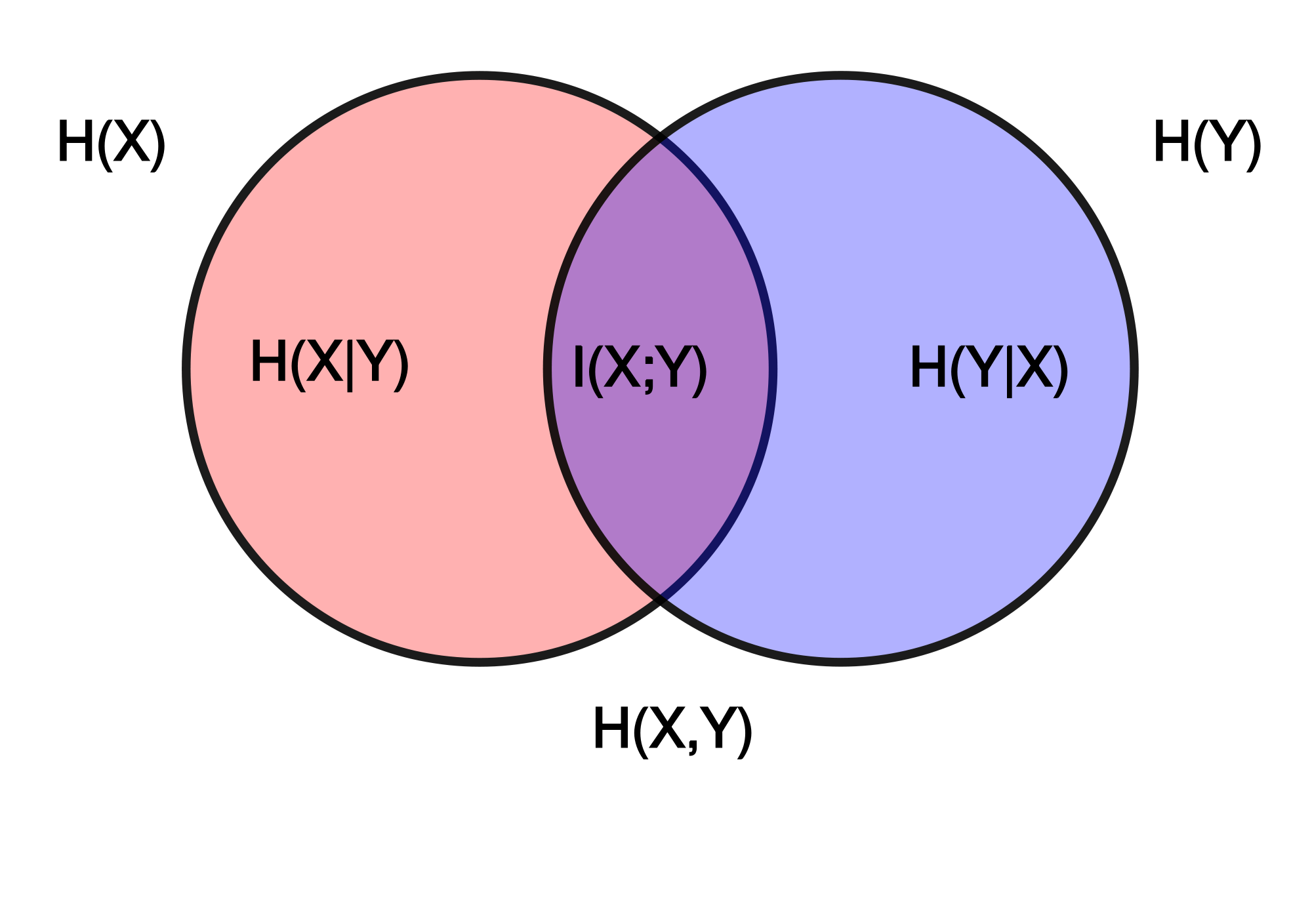
During the bonding period, very interesting discussions came across related to the measures we could develop during the project. As an initial exercise, I thought it could be interesting to test some basic Information Theory based measures. Basically, I wanted to reproduce the calculation of Entropy and the related measures shown in this image:
The calculation of the Entropy is quite straightforward, so I decided to test different implementations of some people and test them with the timeit module.
It turned that among 4-5 different functions I found this one coded using numpy and allowing also the calculation of the Joint Entropy using itertools was the fastest by far.
1 2 3 4 5 6 7 8 9 10 | |
Having the basic blocks, I built the Conditional Entropy and Mutual Information easily:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 | |
I proved the functions over small vectors and all the properties seemed to work well. In the other hand, I sorted out the loading of the fMRI files (that I was loading with an external function) and the calculation of the TR using just the NIFTI header using nibabel.
The first time trying to calculate the Entropy voxel-wise (it took like 5 mins and used only 1 core, so this must be solved in some way or another) and over 90 timeseries extracted from a ROI-mask, I failed. As these measures are calculated over probability density functions, the data must be converted into ‘states’, binarized for example. This is another problem I have to solve: Dealing with fMRI datatypes and FSL functions. I still don’t know how to make use directly of FSL utils (and neither about the different FSL options I have. This is a TO DO for my list).
To solve the problem, I’ve created my own timeseries extractor (generates a Numpy array, not a CSV file as in gen_roi_timeseries.py):
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 | |
This structure can be used to extract all voxels timeseries as well. As work for the next week, this data has to be binarized to two (or more) ‘states’ and then passed to the IT functions.