In this blog I will write about my experience with C-PAC development. Our intention here is to develop a series of scripts that allow us (C-PAC users) to calculate some measures over non-linear fMRI timeseries and integrate them in a workflow. Our aim is to extend and improve C-PAC’s features.
In this first post I will try to explain some of what I have done for my first two scripts for C-PAC as my first contact with C-PAC. I would like to learn a lot about C-PAC’s structure, workflows etc. and give back too. The goal of this first scripts is to say ‘hi’, have some feedback from main developers and take a first hands-on with the code.
In order to learn from C-PAC’s repo on on GitHub and to C-PAC’s forums and I saw a question on cross correlations and though it could be a good exercise to try possibilities (out of using other available utils like FSLcc) and afterwards, I also made the calculation for Transfer Entropy (TE) Granger Causality (GC)). As a short description, these 2 scripts calculate correlations and GC between ROI-time series extracted from an fMRI file (this is done using a ROI-mask with the gen_roi_timeserier.py function which is already in C-PAC).
My code is in the copy I forked, in a new branch called series_mod. I saved my scripts in a folder of the same name.
We have got here 3 files, init _ .py, series_mod.py and utils.py_ . I used these three files from a template of other toolbox in C-PAC. series_mod.py generates the workflows:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 | |
There are 3 input parameters; fMRI series, the ROI-mask and TR of the fMRI. This function makes use of the functions in utils.py. Let’s see part of the code (present in my repo):
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 | |
It is a trivial computation after extracting the time-series. This data could have been treated or filtered, but this first scripts were just to train myself with the repo, python, nypipe workflows etc. .
I started from my own implementations in python, but also took some of this code and ideas from these tutorials:
[1] http://nipy.org/nitime/examples/resting_state_fmri.html
[2] http://nipy.org/nitime/examples/granger_fmri.html
The result for the correlation when making calculations on preprocessed data of 3mm voxel-size and a mask of 90 ROIs parcellation is
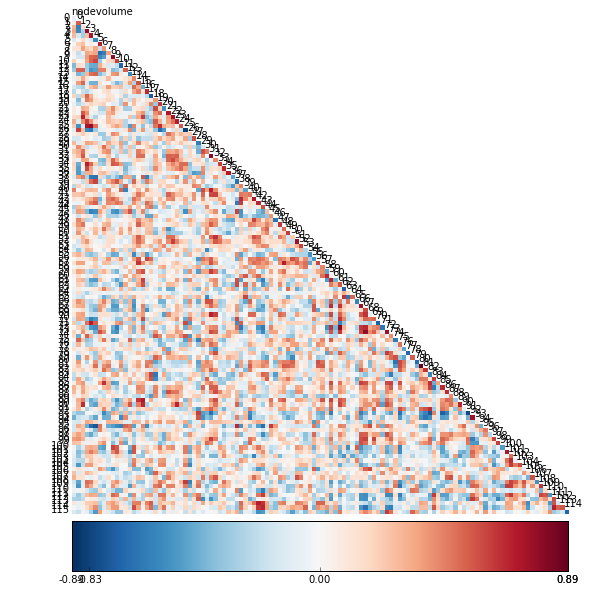
I will discuss about the measures we are going to work on in the next post.